 19459003]data-original-mos=”https://cdn.mos.cms.futurecdn.net/kafFKEzebt3WC3X55HKjdT.jpg” data-pin-media=”https://cdn.mos.cms.futurecdn.net/kafFKEzebt3WC3X55HKjdT.jpg” data-pin-nopin=”true”fetchpriority=”high”>
19459003]data-original-mos=”https://cdn.mos.cms.futurecdn.net/kafFKEzebt3WC3X55HKjdT.jpg” data-pin-media=”https://cdn.mos.cms.futurecdn.net/kafFKEzebt3WC3X55HKjdT.jpg” data-pin-nopin=”true”fetchpriority=”high”>
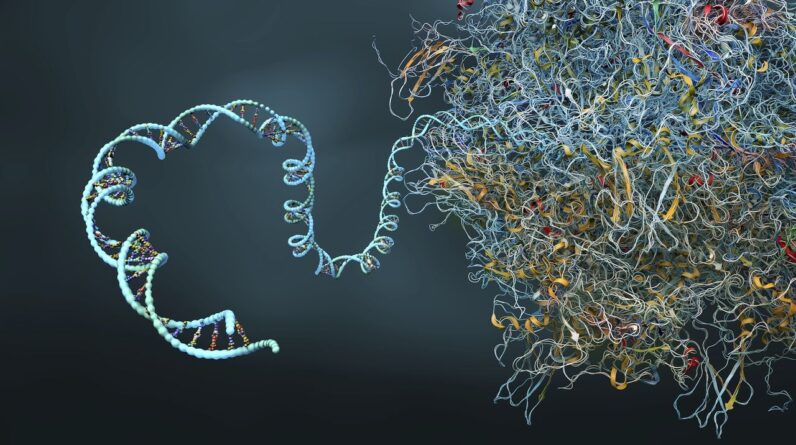
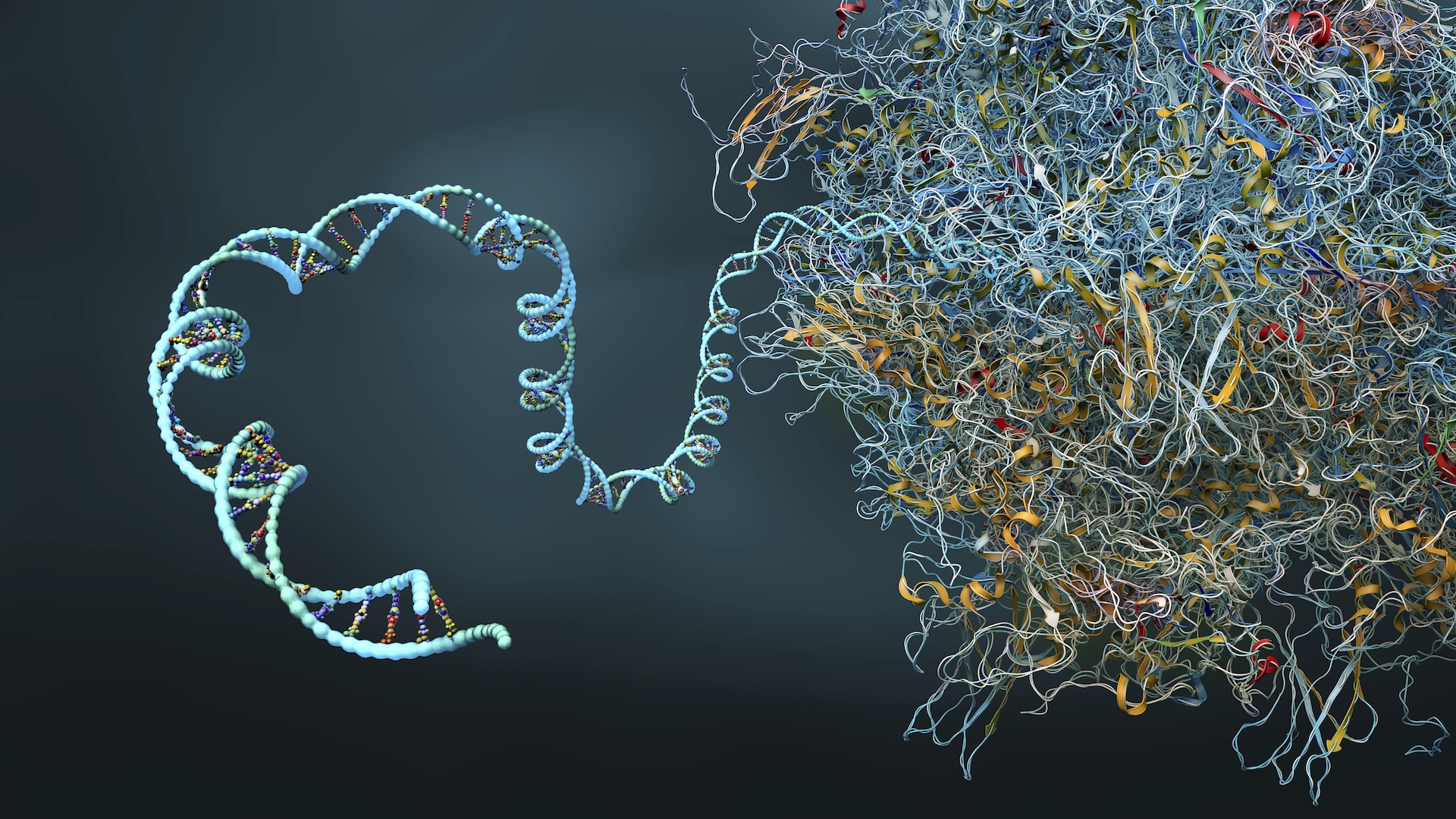
Scientists at IBM and Moderna have actually effectively utilized a quantum simulation algorithm to anticipate the complex secondary protein structure of a 60-nucleotide-long mRNA series, the longest ever simulated on a quantum computer system
Messenger ribonucleic acid (mRNA)is a particle that brings hereditary info from DNA to ribosomes. It directs protein synthesis in cells and is utilized to develop efficient vaccines efficient in prompting particular immune actions.
It’s extensively thought that all the info needed for a protein to embrace the appropriate three-dimensional conformation is supplied by its amino acid series or “folding.”It’s made up of just a single hair of amino acids, mRNA has a secondary protein structure consisting of a series of folds that offer an offered particle’s particular 3D shape. The variety of possible folding permutations increases greatly with each included nucleotide. This makes the difficulty of anticipating what form a mRNA particle will take intractable at greater scales.
The IBM and Moderna experiment, described in a research study Released for the 2024 IEEE International Conference on Quantum Computing and Engineering, showed how quantum computing can be utilized to enhance the conventional techniques for making such forecasts. Generally, these forecasts generally depended on binary, classical computer systems and expert system (AI) designs such as Google DeepMind’s AlphaFold
Related: DeepMind’s AI program AlphaFold3 can anticipate the structure of every protein in deep space– and demonstrate how they work
According to a brand-new research study released May 9 on the preprint arXiv database, algorithms efficient in operating on these classical architectures can process mRNA series with “hundreds or thousands of nucleotides,” Just by omitting greater intricacy functions such as “pseudoknots.”
Get the world’s most interesting discoveries provided directly to your inbox.
Pseudoknots are made complex twists and shapes in a particle’s secondary structure that can participating in more complicated internal interactions than regular folds. Through their exemption, the possible precision of any protein-folding forecast design is essentially restricted.
Comprehending and forecasting even the tiniest information of a mRNA particle’s protein folds is intrinsic to establishing more powerful forecasts and, as an outcome, more efficient mRNA-based vaccines
Researchers want to get rid of the constraints fundamental in the most effective supercomputers and AI designs by enhancing explores quantum innovation. The scientists carried out several experiments utilizing quantum simulation algorithms that count on qubits — the quantum equivalent of a computer system bit– to design particles.
Utilizing just 80 qubits (out of a possible 156) on the R2 Heron quantum processing system (QPU),, the group utilized a conditional value-at-risk-based variational quantum algorithm (CVaR-based VQA)– a quantum optimization algorithm imitated particular methods utilized to evaluate intricate interactions such as accident avoidance and monetary danger evaluation methods — to anticipate the secondary protein structure of a 60-nucleotide-long mRNA series.
The previous finest for a quantum-based simulation design, according to the research studywas a 42-nucleotide series. The scientists likewise scaled the experiment by using current error-correction strategies to handle the sound created by quantum functions
In the brand-new preprint research study, the group provisionally showed the speculative paradigm’s efficiency in running simulated circumstances with as much as 156 qubits for mRNA series of as much as 60 nucleotides. They likewise carried out initial research study showing the prospective to use approximately 354 qubits for the very same algorithms in soundless settings.
Seemingly, increasing the variety of qubits utilized to run the algorithm, while scaling the algorithms for extra subroutines, ought to cause more precise simulations and the capability to forecast longer series, they stated.
They kept in mind, nevertheless, that “these approaches require the advancement of sophisticated strategies for embedding these problem-specific circuits into the existing quantum hardware,”– suggesting that much better algorithms and processing architectures will be required to advance the research study.
Tristan is a U.S-based science and innovation reporter. He covers expert system (AI), theoretical physics, and innovative innovation stories.
His work has actually been released in various outlets consisting of Mother Jones, The Stack, The Next Web, and Undark Magazine.
Prior to journalism, Tristan served in the United States Navy for 10 years as a developer and engineer. When he isn’t composing, he takes pleasure in video gaming with his spouse and studying military history.
Find out more
As an Amazon Associate I earn from qualifying purchases.